Installation
Download and install Amazon Genomics CLI
Download the Amazon Genomics CLI zip, unzip its contents, and run the install.sh script:
To download a specific release, see releases page of our Github repo.
To download the latest release navigate to https://github.com/aws/amazon-genomics-cli/releases/
Once you have downloaded a release, type the following to install:
The latest nightly build can be accessed here: s3://healthai-public-assets-us-east-1/amazon-genomics-cli/nightly-build/amazon-genomics-cli.zip
You can download the nightly by running the following:
aws s3api get-object --bucket healthai-public-assets-us-east-1 --key amazon-genomics-cli/nightly-build/amazon-genomics-cli.zip amazon-genomics-cli.zip
unzip amazon-genomics-cli-<version>.zip
cd amazon-genomics-cli/
./install.sh
This will place the agc command in $HOME/bin.
The Amazon Genomics CLI is a statically compiled Go binary. It should run in your environment natively without any additional setup. Test the CLI with:
$ agc --help
🧬 Launch and manage genomics workloads on AWS.
Commands
Getting Started 🌱
account Commands for AWS account setup.
Install or remove AGC from your account.
Contexts
context Commands for contexts.
Contexts specify workflow engines and computational fleets to use when running a workflow.
Logs
logs Commands for various logs.
Projects
project Commands to interact with projects.
Workflows
workflow Commands for workflows.
Workflows are potentially-dynamic graphs of computational tasks to execute.
Settings ⚙️
configure Commands for configuration.
Configuration is stored per user.
Flags
--format string Format option for output. Valid options are: text, table, json (default "text")
-h, --help help for agc
--silent Suppresses all diagnostic information.
-v, --verbose Display verbose diagnostic information.
--version version for agc
Examples
Displays the help menu for the specified sub-command.
`$ agc account --help`
If this doesn’t work immediately, try:
- start a new terminal shell
- modifying your
$HOME/.bashrc(or equivalent file) appending the following line and restarting your shell:
export PATH=$HOME/bin:$PATH
If you are running this on MacOS, you may see this below popup window when you initially run any agc commands due to Apple’s security restrictions.
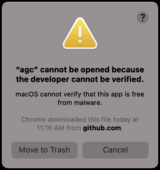
Click Cancel and navigate to Apple’s System Preferences, click Security & Privacy, then click General. Near the bottom, you will see a line indicating "agc" was blocked from use because it is not from an identified developer. To the right, click Allow Anyway.
Now go back to the terminal and run agc --help again. You will see this new popup window below asking you to override the system security.
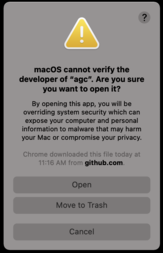
Click Open and now your agc is correctly installed.
Verify that you have the latest version of Amazon Genomics CLI with:
agc --version
If you do not, you may need to uninstall any previous versions of Amazon Genomics CLI and reinstall the latest.
Command Completion
Amazon Genomics CLI can generate shell completion scripts that enable ‘Tab’ completion of commands. Command completion is optional and not required to use Amazon Genomics CLI. To generate a completion script you can use:
agc completion <shell>
where “shell” is one of:
Bash
source <(agc completion bash)
To load completions for each session, execute once:
Linux:
agc completion bash > /etc/bash_completion.d/agc
macOS:
If you haven’t already installed bash-completion, execute the following once
brew install bash-completion
and then, add the following line to your ~/.bash_profile:
[[ -r "/usr/local/etc/profile.d/bash_completion.sh" ]] && . "/usr/local/etc/profile.d/bash_completion.sh"
Once bash completion is installed
agc completion bash > /usr/local/etc/bash_completion.d/agc
Zsh:
If shell completion is not already enabled in your environment, you will need to enable it. You can execute the following once:
echo "autoload -U compinit; compinit" >> ~/.zshrc
To load completions for each session, execute once:
agc completion zsh > "${fpath[1]}/_agc"
You will need to start a new shell for this setup to take effect.
fish:
agc completion fish | source
To load completions for each session, execute once:
agc completion fish > ~/.config/fish/completions/agc.fish
PowerShell:
agc completion powershell | Out-String | Invoke-Expression
To load completions for every new session, run:
agc completion powershell > agc.ps1
and source this file from your PowerShell profile.