Cromwell
Description
Cromwell is a workflow engine developed by the Broad Institute. In Amazon Genomics CLI, Cromwell is an engine that can be deployed in a context as an engine to run workflows based on the WDL specification.
Cromwell is an open source project distributed by the Broad Institute under the Apache 2 license and available on GitHub.
Customizations
Some minor customizations where made to the AWS Backend adapter for Cromwell to facilitate improved scalability and cross region S3 bucket access when deployed with Amazon Genomics CLI. The fork containing these customizations is available here and we are working to contribute these back to the main code base.
Architecture
There are four components of a Cromwell engine as deployed in an Amazon Genomics CLI context.
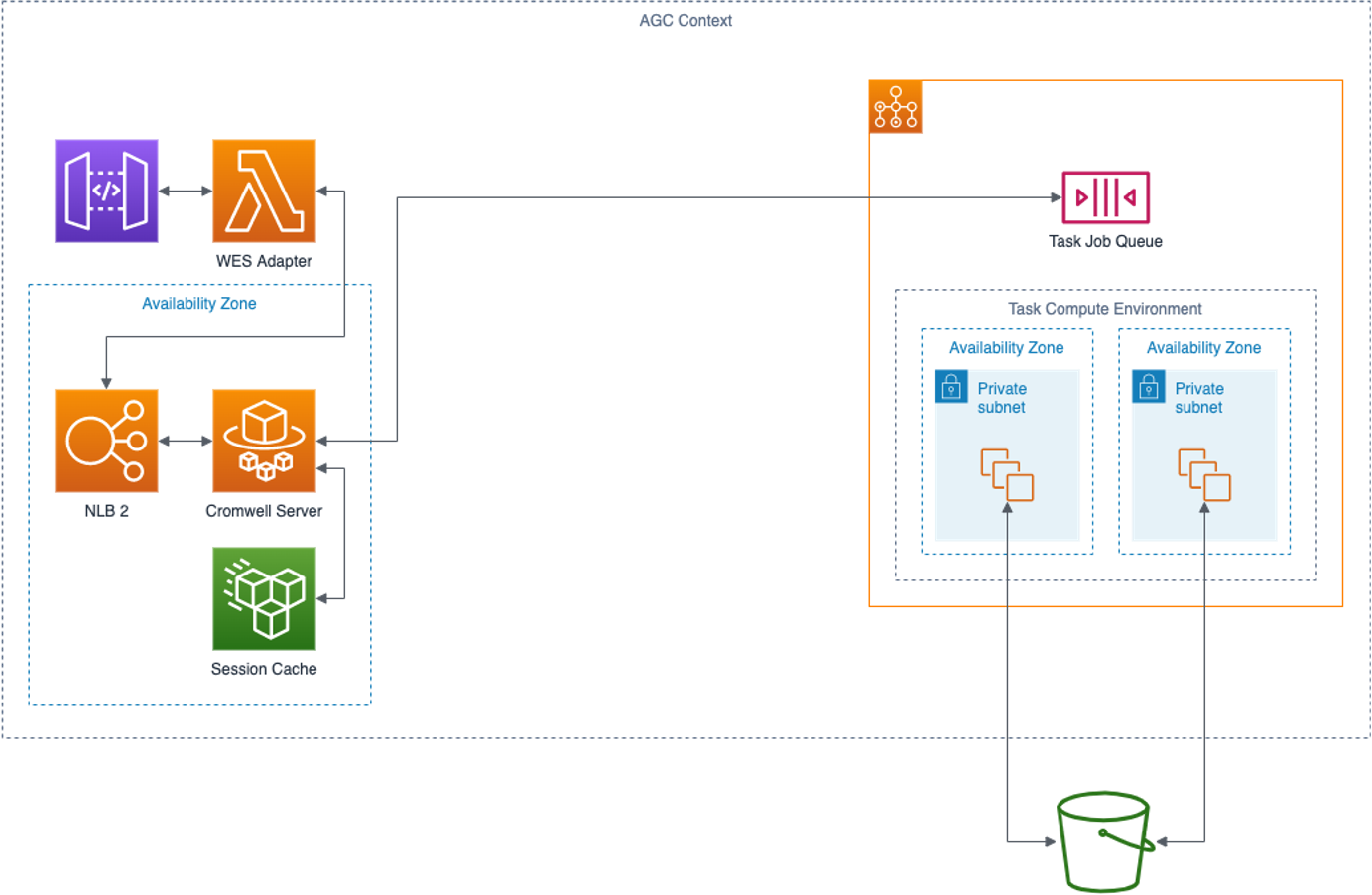
WES Adapter
Amazon Genomics CLI communicates with the Cromwell engine via a GA4GH WES REST service. The WES Adapter implements the WES standard and translates WES calls into calls to the Cromwell REST API. The adapter runs as an Amazon ECS service available via API Gateway.
Cromwell Server
The Cromwell engine is run in “server mode” as a container service in ECS and receives instructions from the WES Adapter. The engine can run multiple workflows asynchronously. Workflow tasks are run in an elastic compute environment and monitored by Cromwell.
Session Cache
Cromwell can use workflow run metadata to perform call caching. When deployed by Amazon Genomics CLI call caching is enabled by default. Metadata is stored by an embedded HSQL DB with file storage in an attached EFS volume. The EFS volume exists for the lifetime of the context the engine is deployed in so re-runs of workflows within the lifetime can benefit from call caching.
Task Compute Environment
Workflow tasks are submitted by Cromwell to an AWS Batch queue and run in containers using an AWS Compute Environment.
Container characteristics are defined by the runtime. AWS Batch coordinates the elastic provisioning of EC2 instances (container hosts)
based on the available work in the queue. Batch will place containers on container hosts as space allows.
Fetch and Run Strategy
Execution of workflow tasks uses a “Fetch and Run” strategy. The commands specified in the command section of the WDL task
are written as a file to S3 and “fetched” into the container and run.
The script is “decorated” with instructions to fetch any File inputs from S3 and to write any File outputs back to S3.
Disk Expansion
Container hosts in the Batch compute environment use EBS volumes as local scratch space. As an EBS volume approaches a
capacity threshold, new EBS volumes will be attached and merged into the file system. These volumes are destroyed when
AWS Batch terminates the container host. For this reason it is not necessary to specify disk requirements for the task
runtime and these WDL directives will be ignored.
AWS Batch Retries
The Cromwell AWS Batch backend supports AWS Batch’s task retry option allowing failed tasks to attempt to run again. This
can be useful for adding resilience to a workflow from sporadic infrastructure failures. It is especially useful when using
an Amazon Genomics CLI “spot” context as spot instances can be terminated with minimal warning. To enable retries, add
the following option to your runtime section of a task:
runtime {
...
awsBatchRetryAttempts: <int>
...
}
where <int> is an integer specifying the number of retries up to a maximum of 10.
Although similar to the WDL preemptible option, awsBatchRetryAttempts has differences in how retries are implemented. Notably,
the implementation falls back on the AWS Batch retry strategy and will retry a task that fails for any reason; whereas the preemptible
option is more specific to failures caused by preemption. At this time the preemptible option is not supported by Amazon Genomics CLI
and is ignored.